oxidative phosporylation
Oxidative phosphorylation is the ultimate metabolic pathway of cellular respiration, and follows glycolysis and the Krebs cycle. The enzymes of oxidative phosphorylation are membrane-bound – at the plasma membrane of prokaryotes, and the analogous inner mitochondrial membrane of eukaryotes.
Energy sources such as glucose are initially metabolized (glycolysis) in the cytoplasm and the products are imported into mitochondria, which continue catabolic metabolism employing metabolic pathways that include the Krebs cycle (citric acid cycle), beta-oxidation of fatty acids, and oxidation of amino acids.
Tables Enzymes Functions Krebs cycle Enzymes Cofactors of Krebs Cycle Electron Transport vs Oxidative Phosphorylation :
In oxidative phosphorylation, the energy carrier molecules generated by the citric acid cycle (NADH & FADH2) enter an electron transfer chain that generates a proton gradient by pumping protons (H+) across the membrane.
Electrons from the donors are passed through an electron transport chain to O2, which is reduced to H2O. This multi-step redox process occurs at the mitochondrial inner membrane. The enzymes that catalyze these reactions simultaneously create a proton gradient across the membrane, producing a thermodynamic high-energy state with the potential to do work. Although electron transport occurs with great efficiency, a small percentage of electrons are prematurely leaked to oxygen, resulting in the formation of the toxic free radical, superoxide.
Intracellular mitochondria are remarkably similar to free-living bacteria. The known structural, functional and DNA similarities between mitochondria and bacteria provide strong evidence that mitochondria evolved from intracellular prokaryotic symbionts that took up residence in primitive eukaryotic cells.
Table Enzymes Functions Krebs cycle Electron Transport Chain vs Oxidative Phosphorylation :
Oxidative phosphorylation metabolic pathway: diagram : Brooks-Cole animation oxidative phosphorylation : Boyer animation oxidative phosphorylation : quick animation ~ electron transport chain in mitochondria : quick animation ~ ATP synthesis in mitochondria : quick animation ~ proton gradient in mitochondria : diagram pseudorotation ATP : animation pseudorotation ATP :
In more detail:
1. Complex I (NADH:ubiquinone oxidoreductase) comprising NADH dehydrogenase and ubiquinone transfers two electrons from NADH to the lipid-soluble carrier, ubiquinone (Q). The reduced product of this redox reaction is ubiquinol (QH2), which is free to diffuse within the membrane. Simultaneous with the reduction of ubiquinone, Complex I moves four protons (H+) across the membrane, generating a proton gradient. Complex I is one of the main sites of production of superoxide free radicals, in which premature electron leakage to oxygen occurs.
The pathway of electrons:
NADH is oxidized to NAD+, reducing FMN to FMNH2 in a single two-electron step. The subsequent electron carrier is a Fe-S cluster, which accepts a single electron to reduce the ferric ion into a ferrous ion. FMNH2 can only be oxidized in two one-electron steps, through a semiquinone intermediate. The electron thus travels from the FMNH2 to the Fe-S cluster, and then from the Fe-S cluster to the oxidized Q to generate the free-radical (semiquinone) form of Q. This reduces the semiquinone form to the ubiquinol form, QH2. Simultaneously, four protons are translocated from the matrix across the inner mitochondrial membrane to the intermembrane space. This generates the proton gradient that will be subsequently used to generate ATP through oxidative phosphorylation.
2. Complex II comprises succinate dehydrogenase, which is not a proton pump. It functions to funnel additional electrons into the quinone pool (Q) by removing electrons from succinate and transferring them (via FAD) to Q. Other electron donors such as fatty acids and glycerol 3-phosphate also funnel electrons into Q (via FAD).
3. Complex III comprises cytochrome bc1 complex and removes two electrons from QH2 and stepwise transfers them to two molecules of cytochrome c, which is a water-soluble electron carrier located on the outer surface of the membrane. Simultaneously, Complex III pumps four protons across the membrane, generating a proton gradient. When electron transfer is hindered (by a high membrane potential, point mutations, or by respiratory inhibitors such as antimycin A), Complex III may leak electrons to O2 resulting in the formation of a superoxide.
4. Complex IV comprises cytochrome c oxidase, which removes four electrons from four molecules of cytochrome c and transfers them to molecular oxygen (O2), producing two molecules of water (H2O). Simultaneously, it pumps four protons across the membrane, generating a proton gradient.
Thus, the electron transport chain comprises four complexes, of which three complexes (I, III, IV) pump protons across the inne mitochondrial membrane to generate a proton gradient that supplies energy to ATP synthase, which generates ATP in oxidative phosphorylation. ATP synthase is sometimes regarded as complex V of the electron transport chain, though strictly it merely follows the chain.
ATP synthase enzymes have been considerably conserved through evolution. The enzymes in bacteria are essentially the same in structure and function as those in the mitochondria of animals, plants and fungi, and the chloroplasts of plants. There are minor differences between bacteria, mitochondria and chloroplasts in some of the smaller subunits, leading to a confusing nomenclature. The simplest ATP sunthase system is that in E. coli.
The early ancestory of the enzyme is observable in the fact that Archaea have an enzyme that is clearly closely related, yet exhibits significant differences from the Eubacterial branch. The H+-ATP-ase found in vacuoles in the cell cytoplasm of eukaryotes is similar to the archaeal enzyme, and this is thought to reflect its origin from an archaeal ancestor.
In most systems, the ATP synthase is embedded in the membrane (the "coupling" membrane), and catalyses the synthesis of ATP from ADP and phosphate, which is driven by a flux of protons across the membrane and down the proton gradient generated by electron transfer. The flux flows from the protochemically positive (P) side with a high proton electrochemical potential to the protochemically negative (N) side. The reaction catalyzed by ATP synthase is fully reversible, so ATP hydrolysis generates a proton gradient by a reversal of this flux. In some bacteria, the chief function of ATP synthase moves in the direction of ATP hydrolysis, employing ATP generated by fermentative metabolism to provide a proton gradient that drives substrate accumulation, and maintains ionic balance.
ADP + Pi + nH+P <=> ATP + nH+N
View ATP Synthase Animation : Animation movies of ATP synthase : Yoshida Hisabori movies : Nature animation ATP synthase : Pictures & Movies : ATP synthesis ~ animation : animation of ATP synthesis :
Table Electron Transport Chain vs Oxidative Phosphorylation Enzymes Functions Krebs cycle :
The reactions catalyzed by Complex I and Complex III operate close to equilibrium, such that the steady-state concentrations of the reactants and products are approximately equal. Thus, these reactions are readily reversible by increasing the concentration of the products relative to the concentration of the reactants (for example, by increasing the proton gradient).
ATP synthase is also readily reversible.
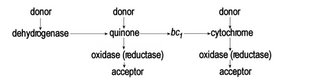
Thus, ATP can be used to generate a proton gradient, which in turn can be employed to generate NADH, which is the reverse of the eukaryotic mechanism that employs NADH as the chief electron donor for the ultimate generation of ATP by oxidative phosphorylation. This process of reverse electron transport is important in many prokaryotic electron transport chains, in which there exist a variety of electron donors and electron acceptors – a variety of dehydrogenases, of oxidases, of reductases, and of electron acceptors:
Energy sources such as glucose are initially metabolized (glycolysis) in the cytoplasm and the products are imported into mitochondria, which continue catabolic metabolism employing metabolic pathways that include the Krebs cycle (citric acid cycle), beta-oxidation of fatty acids, and oxidation of amino acids.
Tables Enzymes Functions Krebs cycle Enzymes Cofactors of Krebs Cycle Electron Transport vs Oxidative Phosphorylation :
In oxidative phosphorylation, the energy carrier molecules generated by the citric acid cycle (NADH & FADH2) enter an electron transfer chain that generates a proton gradient by pumping protons (H+) across the membrane.
Electrons from the donors are passed through an electron transport chain to O2, which is reduced to H2O. This multi-step redox process occurs at the mitochondrial inner membrane. The enzymes that catalyze these reactions simultaneously create a proton gradient across the membrane, producing a thermodynamic high-energy state with the potential to do work. Although electron transport occurs with great efficiency, a small percentage of electrons are prematurely leaked to oxygen, resulting in the formation of the toxic free radical, superoxide.
Intracellular mitochondria are remarkably similar to free-living bacteria. The known structural, functional and DNA similarities between mitochondria and bacteria provide strong evidence that mitochondria evolved from intracellular prokaryotic symbionts that took up residence in primitive eukaryotic cells.
Table Enzymes Functions Krebs cycle Electron Transport Chain vs Oxidative Phosphorylation :
Oxidative phosphorylation metabolic pathway: diagram : Brooks-Cole animation oxidative phosphorylation : Boyer animation oxidative phosphorylation : quick animation ~ electron transport chain in mitochondria : quick animation ~ ATP synthesis in mitochondria : quick animation ~ proton gradient in mitochondria : diagram pseudorotation ATP : animation pseudorotation ATP :
In more detail:
1. Complex I (NADH:ubiquinone oxidoreductase) comprising NADH dehydrogenase and ubiquinone transfers two electrons from NADH to the lipid-soluble carrier, ubiquinone (Q). The reduced product of this redox reaction is ubiquinol (QH2), which is free to diffuse within the membrane. Simultaneous with the reduction of ubiquinone, Complex I moves four protons (H+) across the membrane, generating a proton gradient. Complex I is one of the main sites of production of superoxide free radicals, in which premature electron leakage to oxygen occurs.
The pathway of electrons:
NADH is oxidized to NAD+, reducing FMN to FMNH2 in a single two-electron step. The subsequent electron carrier is a Fe-S cluster, which accepts a single electron to reduce the ferric ion into a ferrous ion. FMNH2 can only be oxidized in two one-electron steps, through a semiquinone intermediate. The electron thus travels from the FMNH2 to the Fe-S cluster, and then from the Fe-S cluster to the oxidized Q to generate the free-radical (semiquinone) form of Q. This reduces the semiquinone form to the ubiquinol form, QH2. Simultaneously, four protons are translocated from the matrix across the inner mitochondrial membrane to the intermembrane space. This generates the proton gradient that will be subsequently used to generate ATP through oxidative phosphorylation.
2. Complex II comprises succinate dehydrogenase, which is not a proton pump. It functions to funnel additional electrons into the quinone pool (Q) by removing electrons from succinate and transferring them (via FAD) to Q. Other electron donors such as fatty acids and glycerol 3-phosphate also funnel electrons into Q (via FAD).
3. Complex III comprises cytochrome bc1 complex and removes two electrons from QH2 and stepwise transfers them to two molecules of cytochrome c, which is a water-soluble electron carrier located on the outer surface of the membrane. Simultaneously, Complex III pumps four protons across the membrane, generating a proton gradient. When electron transfer is hindered (by a high membrane potential, point mutations, or by respiratory inhibitors such as antimycin A), Complex III may leak electrons to O2 resulting in the formation of a superoxide.
4. Complex IV comprises cytochrome c oxidase, which removes four electrons from four molecules of cytochrome c and transfers them to molecular oxygen (O2), producing two molecules of water (H2O). Simultaneously, it pumps four protons across the membrane, generating a proton gradient.
Thus, the electron transport chain comprises four complexes, of which three complexes (I, III, IV) pump protons across the inne mitochondrial membrane to generate a proton gradient that supplies energy to ATP synthase, which generates ATP in oxidative phosphorylation. ATP synthase is sometimes regarded as complex V of the electron transport chain, though strictly it merely follows the chain.
ATP synthase enzymes have been considerably conserved through evolution. The enzymes in bacteria are essentially the same in structure and function as those in the mitochondria of animals, plants and fungi, and the chloroplasts of plants. There are minor differences between bacteria, mitochondria and chloroplasts in some of the smaller subunits, leading to a confusing nomenclature. The simplest ATP sunthase system is that in E. coli.
The early ancestory of the enzyme is observable in the fact that Archaea have an enzyme that is clearly closely related, yet exhibits significant differences from the Eubacterial branch. The H+-ATP-ase found in vacuoles in the cell cytoplasm of eukaryotes is similar to the archaeal enzyme, and this is thought to reflect its origin from an archaeal ancestor.
In most systems, the ATP synthase is embedded in the membrane (the "coupling" membrane), and catalyses the synthesis of ATP from ADP and phosphate, which is driven by a flux of protons across the membrane and down the proton gradient generated by electron transfer. The flux flows from the protochemically positive (P) side with a high proton electrochemical potential to the protochemically negative (N) side. The reaction catalyzed by ATP synthase is fully reversible, so ATP hydrolysis generates a proton gradient by a reversal of this flux. In some bacteria, the chief function of ATP synthase moves in the direction of ATP hydrolysis, employing ATP generated by fermentative metabolism to provide a proton gradient that drives substrate accumulation, and maintains ionic balance.
ADP + Pi + nH+P <=> ATP + nH+N
View ATP Synthase Animation : Animation movies of ATP synthase : Yoshida Hisabori movies : Nature animation ATP synthase : Pictures & Movies : ATP synthesis ~ animation : animation of ATP synthesis :
Table Electron Transport Chain vs Oxidative Phosphorylation Enzymes Functions Krebs cycle :
The reactions catalyzed by Complex I and Complex III operate close to equilibrium, such that the steady-state concentrations of the reactants and products are approximately equal. Thus, these reactions are readily reversible by increasing the concentration of the products relative to the concentration of the reactants (for example, by increasing the proton gradient).
ATP synthase is also readily reversible.
Thus, ATP can be used to generate a proton gradient, which in turn can be employed to generate NADH, which is the reverse of the eukaryotic mechanism that employs NADH as the chief electron donor for the ultimate generation of ATP by oxidative phosphorylation. This process of reverse electron transport is important in many prokaryotic electron transport chains, in which there exist a variety of electron donors and electron acceptors – a variety of dehydrogenases, of oxidases, of reductases, and of electron acceptors:









































0 Comments:
Post a Comment
<< Home